Evolution and conservation genomics of endangered wildlife
This research line is at the interface of ecology and evolution, employing computational approaches to reconstruct population history and to study basic and applied questions associated with adaptation, behavior, and taxonomy. Research programs feature species of conservation concern, utilize population genetics and phylogenetics to assess species’ natural history, define management units and provide scientific foundation for conservation programs in situ and ex situ.
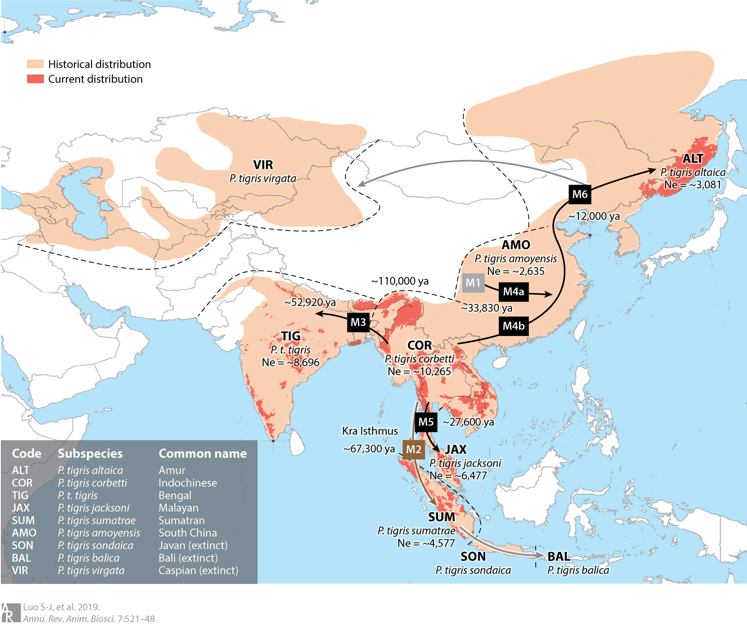
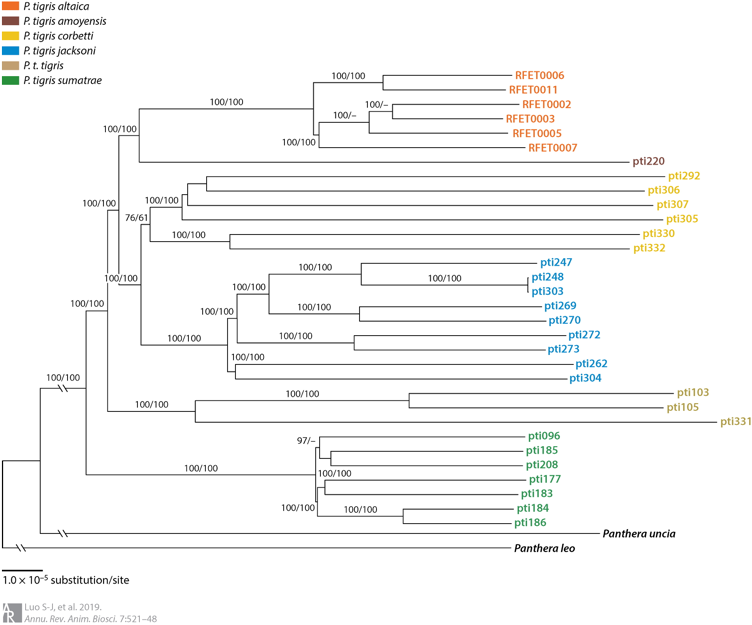
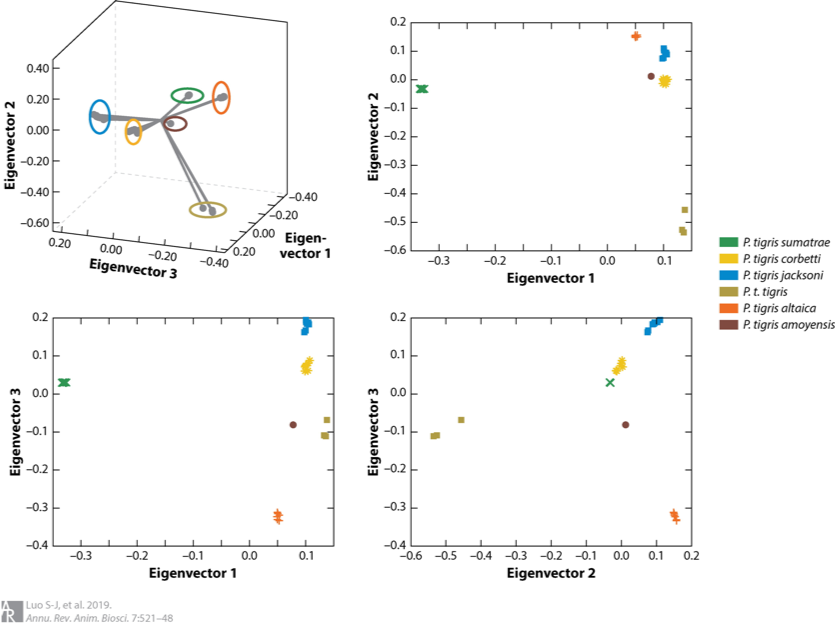
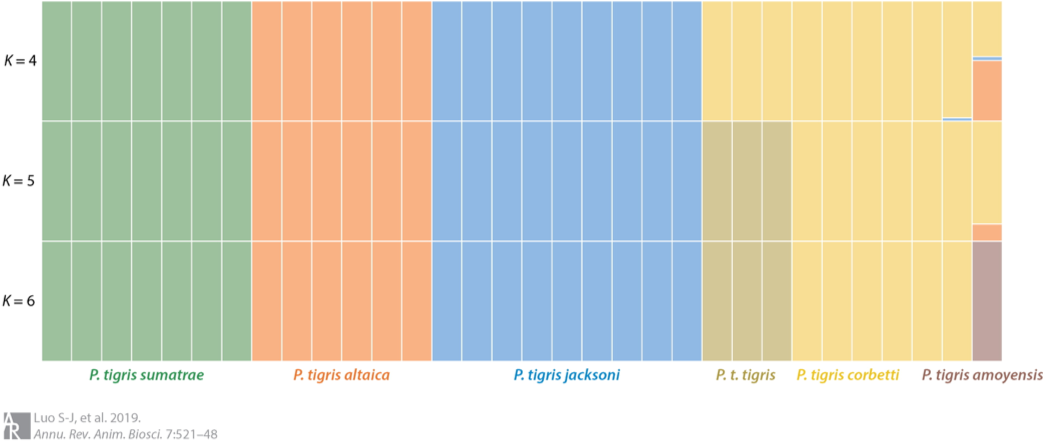
Fewer than 4,000 free-ranging tigers remain in the wild. Efforts to protect these remaining tigers have also been partially stymied by uncertainty about whether they represent six, five or only two subspecies. In 2004, my colleagues and I published our work on tiger genetic ancestry and phylogeography (Luo et al. 2004), in which we recognized six extant subspecies and split the Malayan tiger from its Indochinese counterpart as a distinct, new living subspecies Panthera tigris jacksoni. Subsequently we further elucidated that the extinct Caspian tiger was not significantly different from the extant Amur tiger (Driscoll et al. 2009; Driscoll et al. 2012) and revealed a close molecular genetic affinity of the extinct Javan and Bali tigers and living Sumatran tiger, with ancient DNA approaches (Xue et al. 2015). In addition, some other publications on genetics of tigers including review of conservation genetic applications in tiger (Luo et al. 2010) and optimization of a multiplex microsatellite panel for identification of individual, subspecies and sex in tigers (Zou et al. 2015). More recently, we presented genome-wide evidence for the tiger’s origin, evolution and adaptation based on whole genome sequencing data from 32 voucher specimens (Liu et al. 2018). The time to the most recent common ancestor for modern tigers was traced back to 110 kya and six living subspecies was validated, or Amur, Indochinese, Malayan, Bengal, Sumatran and South China tigers. Signals of positive selection was identified in the Sumatran tiger, likely associated with smaller body size and thus adaptation to the tropical Sunda Islands (Liu et al. 2018; Luo et al. 2019).
In addition, this line of research has also encompassed a wide range of endangered species with evolutionary or conservation significance. We completed comparative phylogeography of six sympatric Asian felids based on 445 specimens and revealed several novel aspects about the biogeographical history of Southeast Asia, including a major divergence between the Indochinese and Sundaic felids on the Thai-Malay Peninsula (Luo et al. 2014). The first complete phylogeny of pangolins was generated, providing a solid scientific basis for molecular tracing of the world’s most trafficked mammals (Gaubert et al. 2017). Within the landscape of China, we illustrated for the first time the very recent fragmentation effect of the Qinghai-Tibet railway on the landscape genetics of the endangered Przewalski’s gazelle (Yu et al. 2017).
- Genetic basis of morphology and adaptation in Felidae and other mammalian species
- Field ecology and conservation of wild felids in Asia